Scaling Probabilistic Models with Variational Inference
PyData Berlin 2025
Outline
Motivating Examples
Variational Inference in a Nutshell
Toy Example: Parameter Recovery Gamma Distribution
End-to-End Example: Bayesian Neural Network Model
Tips & Tricks
References
What is Variational Inference?
Bayes’ Rule
Recall, given the data \(D\) and the model \(p(\theta|D)\), we can use Bayes’ rule to compute the posterior distribution:
\[ p(\theta|D) = \frac{p(D|\theta)p(\theta)}{p(D)} \]
where:
- \(p(D|\theta)\): Likelihood (how well the model fits the data)
- \(p(\theta)\): Prior (our beliefs about the parameters before seeing the data)
- \(\displaystyle {p(D) = \int p(D|\theta)p(\theta)d\theta}\): Hard to compute!
Stochastic Variational Inference (SVI) is a scalable approximate inference method that transforms the problem of posterior inference into an optimization problem.
Instead of sampling from the posterior distribution (like MCMC), SVI finds the best approximation to the posterior within a family of simpler distributions.
Variational Inference in PyMC
JAX Ecosystem
JAX is a Python library for accelerator-oriented array computation and program transformation, designed for high-performance numerical computing and large-scale machine learning.
JAX
NumPyro

Flax
Optax
Variational Inference in a Nutshell
\(D = \{(x_i, y_i)\}_{i=1}^N\): Our complete dataset
\(\theta\): Model parameters (e.g., in a neural network, the weights and biases)
\(p(\theta|D)\): True posterior distribution (what we want but can’t compute easily)
\(q_\phi(\theta)\): Variational approximation to the posterior (what we’ll optimize)
\(\phi\): Variational parameters (parameters of our approximate posterior)
\(\text{ELBO}(\phi) = \mathbb{E}_{q_\phi(\theta)}[\log p(y|x, \theta)] - \text{KL}(q_\phi(\theta) || p(\theta))\)
Maximizing the ELBO is equivalent to minimizing the KL divergence between our approximate posterior \(q_\phi(\theta)\) and the true posterior \(p(\theta|D)\) (up to a constant, with respect to \(q_\phi(\theta)\)). Why? Use the Bayes’ rule to expand the KL divergence.
This decomposes into two intuitive terms:
\(\mathbb{E}_{q_\phi(\theta)}[\log p(y|x, \theta)]\): Expected log-likelihood (how well we explain the data)
\(\text{KL}(q_\phi(\theta) || p(\theta))\): KL divergence between \(q_\phi(\theta)\) and the prior \(p(\theta)\).
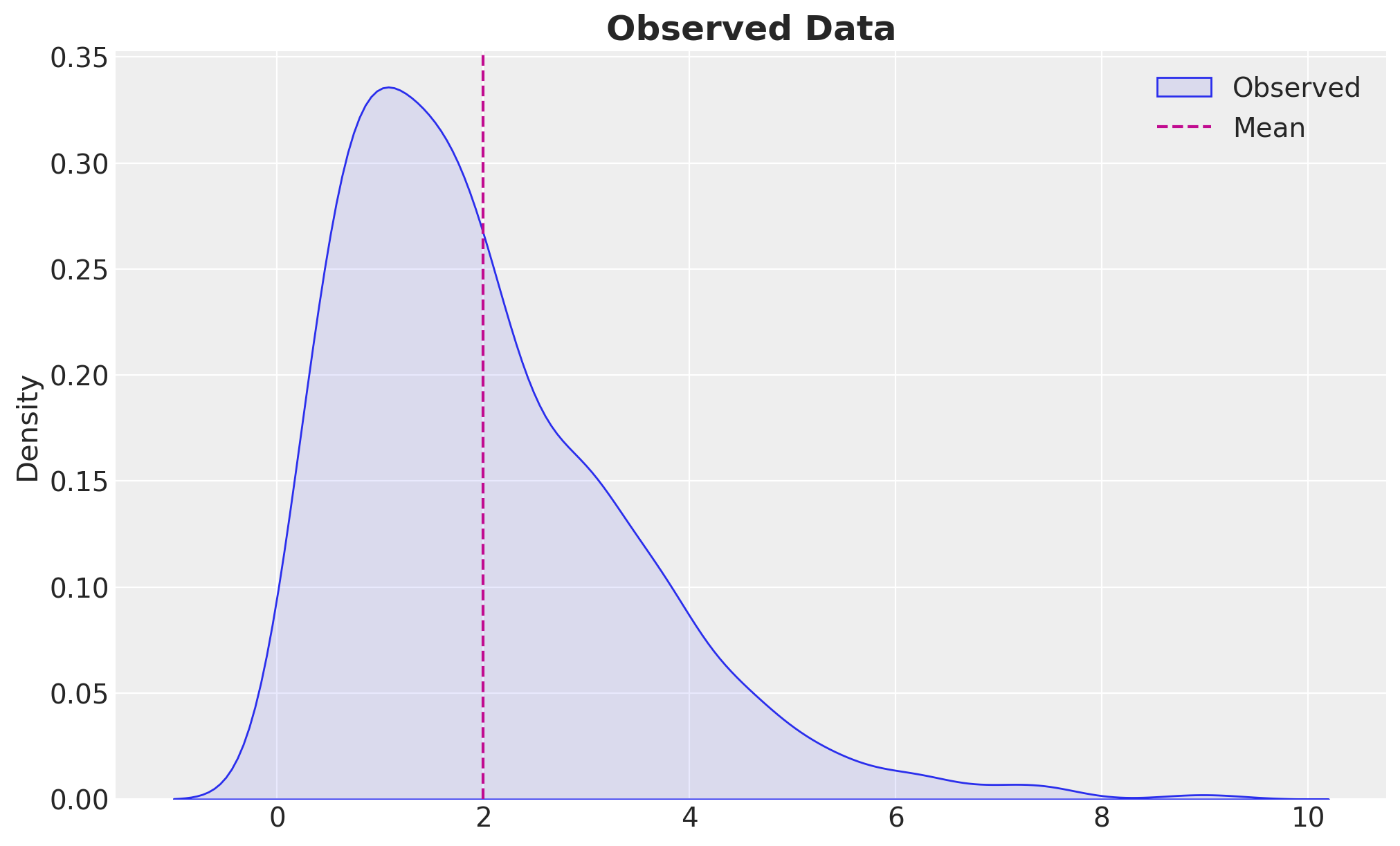
Estimating the Concentration Parameter of a Gamma Distribution \(\text{Gamma}(\alpha) \sim x^{\alpha - 1} e^{-x}\)
Approximation Model
We want to estimate the concentration parameter of a Gamma distribution. We model the data generating process as:
# Model to estimate the concentration parameter
def model(z: jax.Array | None = None) -> None:
# Sample the concentration parameter from a HalfNormal distribution
# as the concentration parameter has to be positive
concentration = numpyro.sample(
"concentration",
dist.HalfNormal(scale=1),
)
# Sample the data from a Gamma distribution
# with the concentration parameter and a rate of 1.0
rate = 1.0
numpyro.sample(
"z",
dist.Gamma(concentration=concentration, rate=rate),
obs=z,
)Approximation Guide
def guide(z: jax.Array | None = None) -> None:
# Location and scale parameters of the normal distribution
concentration_loc = numpyro.param(
"concentration_loc",
init_value=0.5,
)
concentration_scale = numpyro.param(
"concentration_scale",
init_value=0.1,
constraint=dist.constraints.positive,
)
# Variational approximation
base_distribution = dist.Normal(
loc=concentration_loc,
scale=concentration_scale,
)
# We need to transform the base distribution
# as the concentration parameter has to be positive
transformed_distribution = dist.TransformedDistribution(
base_distribution=base_distribution,
transforms=dist.transforms.ExpTransform(),
)
# Sample the concentration parameter from the transformed distribution
numpyro.sample("concentration", transformed_distribution)Inference with SVI
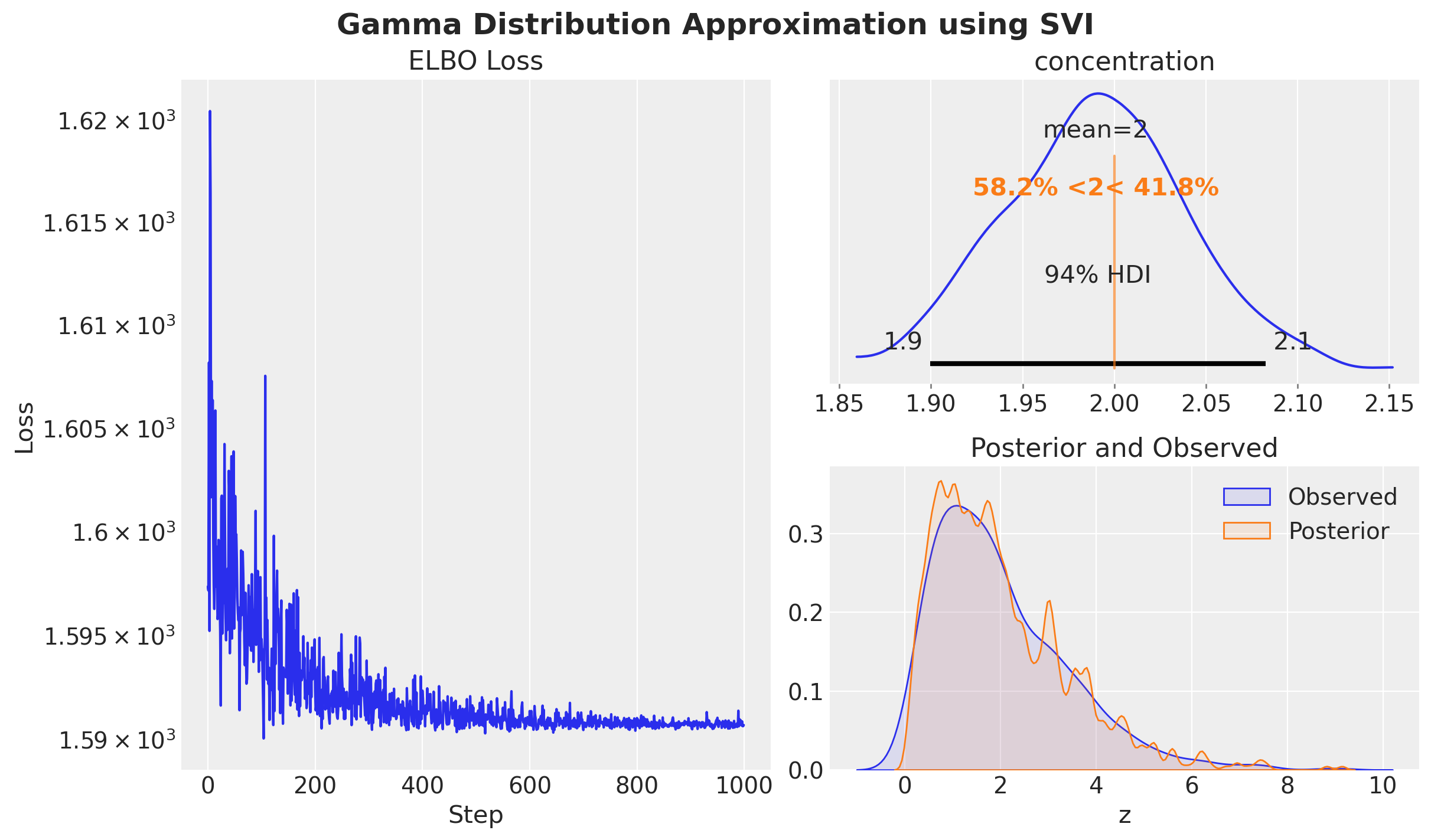
This is how a typical SVI workflow looks like in NumPyro:
# Define the loss function (ELBO)
loss = Trace_ELBO(num_particles=10)
# Define the optimizer
optimizer = optax.adam(learning_rate=0.005)
# Define the SVI algorithm
svi = SVI(model=model, guide=guide, optim=optimizer, loss=loss)
# Run the SVI algorithm
rng_key, rng_subkey = random.split(rng_key)
svi_result = svi.run(
rng_subkey,
num_steps=1_000,
z=z,
progress_bar=True,
)Approximation Results
Example: Bayesian Neural Network
Classification problem with non-linear decision boundary.
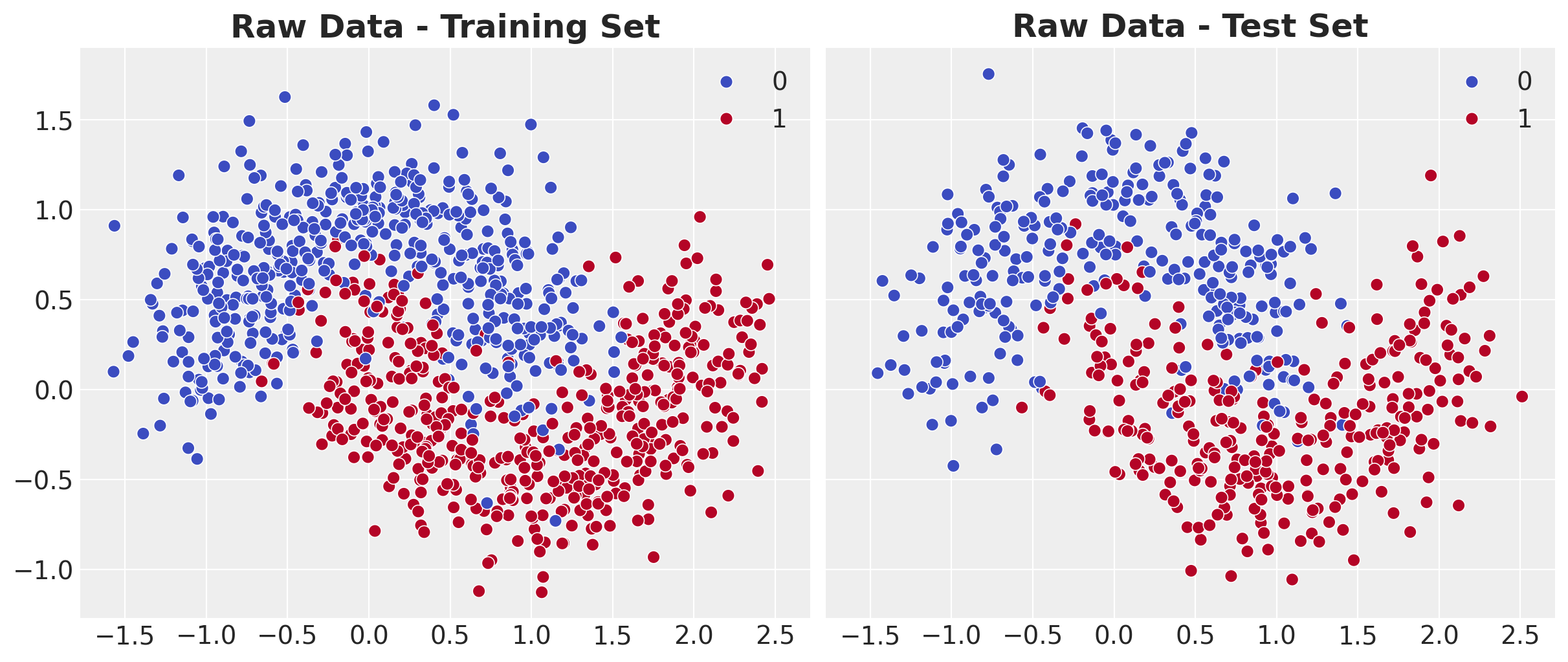
Model Structure
Model Components
- We use a multi-layer perceptron (MLP) to model the non-linear decision boundary parameter \(p(x)\).
- We model our data with a Bernoulli likelihood with parameter \(p(x)\).

Neural Network Component 🤖
import jax
import jax.random as random
from flax import nnx
from itertools import pairwise
class MLP(nnx.Module):
def __init__(
self, din: int, dout: int, hidden_layers: list[int], *, rngs: nnx.Rngs
) -> None:
self.layers = []
layer_dims = [din, *hidden_layers, dout]
for in_dim, out_dim in pairwise(layer_dims):
self.layers.append(nnx.Linear(in_dim, out_dim, rngs=rngs))
def __call__(self, x: jax.Array) -> jax.Array:
for layer in self.layers[:-1]:
x = jax.nn.tanh(layer(x))
return jax.nn.sigmoid(self.layers[-1](x))
hidden_layers = [4, 3]
dout = 1
rng_key, rng_subkey = random.split(rng_key)
nnx_module = MLP(
din=x_train.shape[1],
dout=dout,
hidden_layers=hidden_layers,
rngs=nnx.Rngs(rng_subkey),
)NumPyro Model
import numpyro
import numpyro.distributions as dist
from numpyro.contrib.module import random_nnx_module
def model(
x: Float[Array, "n_obs features"],
y: Int[Array, " n_obs"] | None = None,
) -> None:
n_obs: int = x.shape[0]
def prior(name, shape):
if "bias" in name:
return dist.Normal(loc=0, scale=1)
return dist.SoftLaplace(loc=0, scale=1)
nn = random_nnx_module(
"nn",
nnx_module,
prior=prior,
)
p = numpyro.deterministic("p", nn(x).squeeze(-1))
with numpyro.plate("data", n_obs):
numpyro.sample("y", dist.Bernoulli(probs=p), obs=y)Prior Predictive Checks
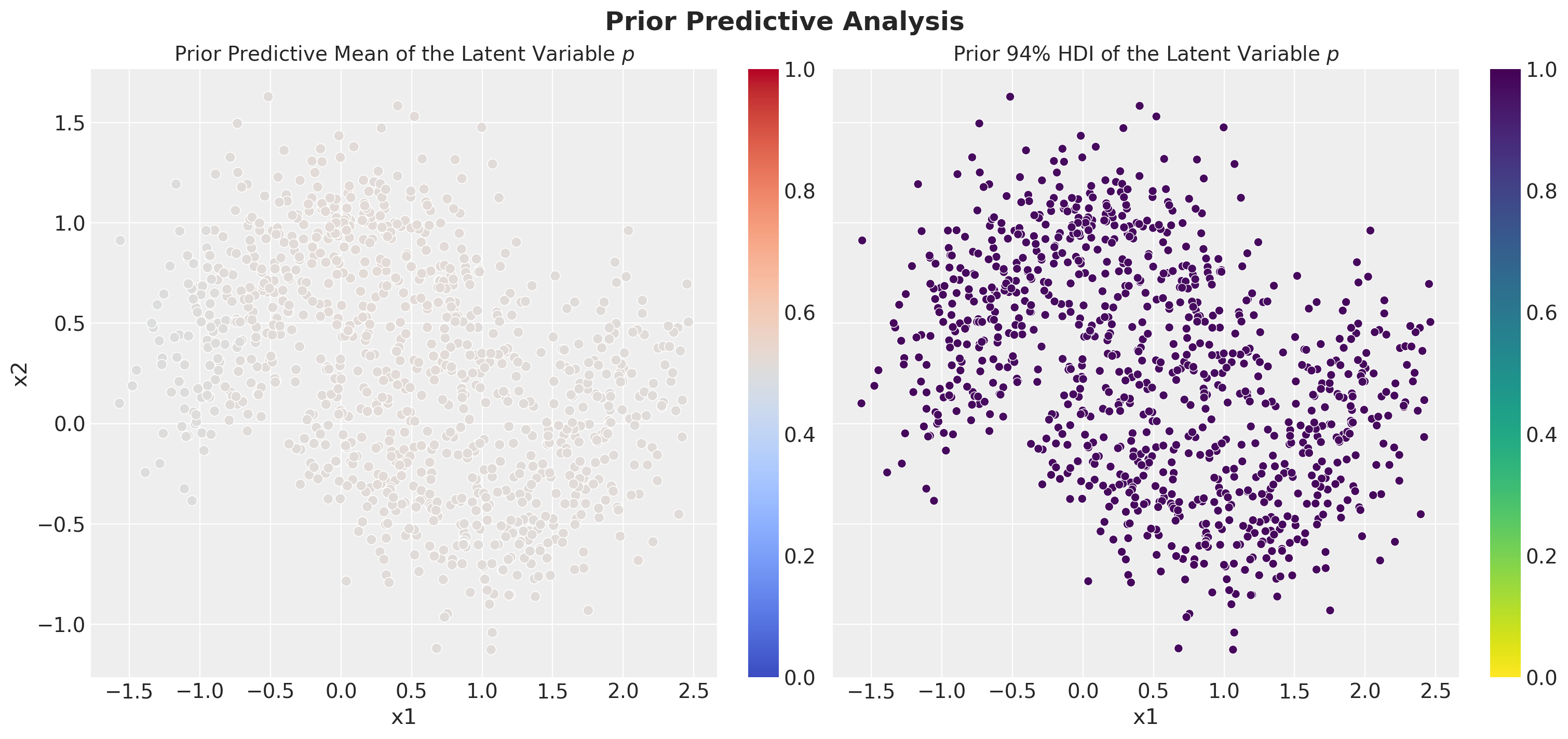
Inference: SVI in NumPyro 🚀
import numpyro
from numpyro.infer import SVI, Trace_ELBO
from numpyro.infer.autoguide import AutoNormal
guide = AutoNormal(model) # <- Posterior Approximation
optimizer = numpyro.optim.Adam(step_size=0.01) # <- Optimizer
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
num_steps = 10_000
rng_key, rng_subkey = random.split(key=rng_key)
# Optimization Loop
svi_result = svi.run(
rng_subkey,
num_steps,
x_train,
y_train,
)
# Results
params = svi_result.paramsInference: ELBO Curve
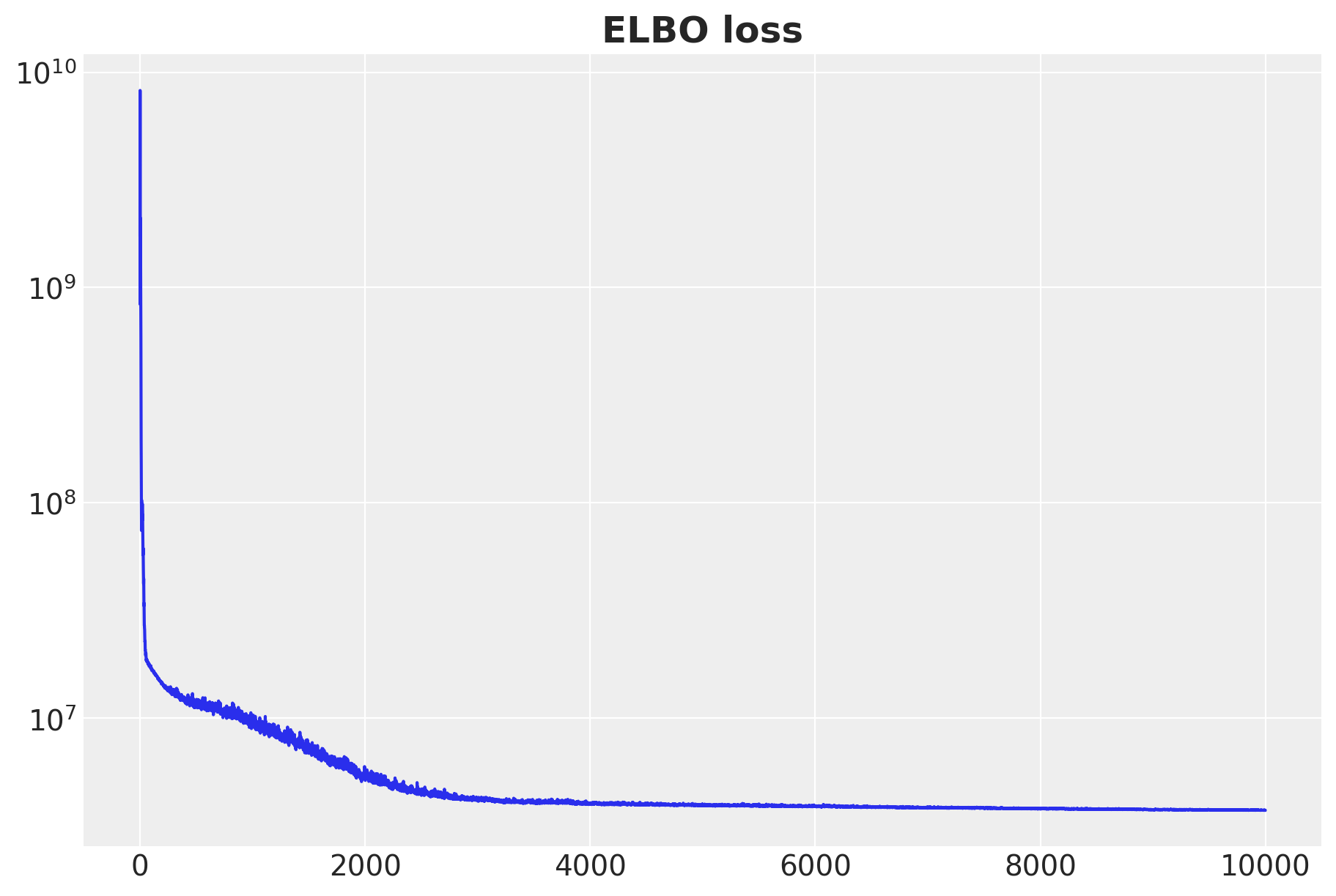
Inference: Custom Guide 🤕
Writing a custom guide?
- If you are having trouble constructing a custom guide, use an
AutoGuide. - Parameter initialization matters: initialize guide distributions to have low variance.
- Pay attention to scales.
Inference: Custom Optimizer ⚡
import optax
# OneCycle schedule: low -> high -> low with specific timing
scheduler = optax.linear_onecycle_schedule(
transition_steps=8_000, # Total number of optimization steps
peak_value=0.008, # Maximum learning rate (reached at pct_start)
pct_start=0.2, # Percent of training to reach peak (20%)
pct_final=0.8, # Percent of training for final phase (80%)
div_factor=3, # Initial LR = peak_value / div_factor
final_div_factor=4, # Final LR = initial_LR / final_div_factor
)
# Chain multiple optimizers for sophisticated training
optimizer = optax.chain(
# Primary optimizer: Adam with scheduled learning rate
optax.adam(learning_rate=scheduler),
# Secondary optimizer: Reduce LR when loss plateaus
optax.contrib.reduce_on_plateau(
factor=0.1, # Multiply LR by 0.1 when plateau detected
patience=10, # Wait 10 evaluations before reducing
accumulation_size=100, # Window size for detecting plateaus
),
)Inference: Custom Optimization Loop
def body_fn(svi_state, _):
svi_state, loss = svi.update(svi_state, x=x_train, y=y_train)
return svi_state, loss
with tqdm.trange(1, num_steps + 1) as t:
batch = max(num_steps // 20, 1)
patience_counter = 0
for i in t:
# Perform one training step (JIT compiled for speed)
svi_state, train_loss = jax.jit(body_fn)(svi_state, None)
# Normalize loss by dataset size for fair comparison
norm_train_loss = jax.device_get(train_loss) / y_train.shape[0]
train_losses.append(jax.device_get(train_loss))
norm_train_losses.append(norm_train_loss)
# Compute validation loss (JIT compiled for speed)
val_loss = jax.jit(get_val_loss)(svi_state, x_val, y_val)
norm_val_loss = jax.device_get(val_loss) / y_val.shape[0]
val_losses.append(jax.device_get(val_loss))
norm_val_losses.append(norm_val_loss)
# Early stopping logic: stop if validation loss > training loss consistently
condition = norm_val_loss > norm_train_loss
patience_counter = patience_counter + 1 if condition else 0
if patience_counter >= patience:
print(
f"Early stopping at step {i} (validation loss exceeding training loss)"
)
breakInference: Early Stopping
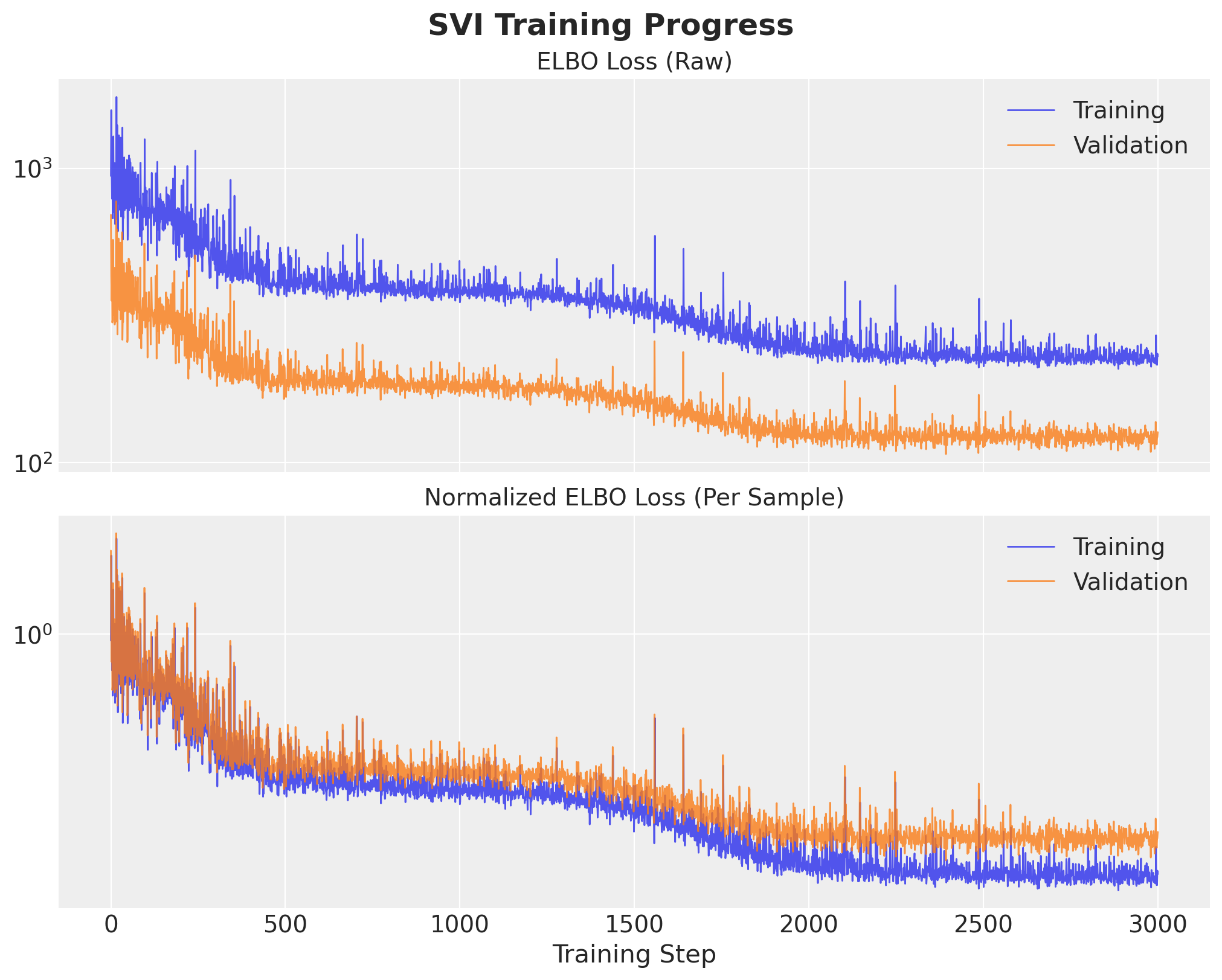
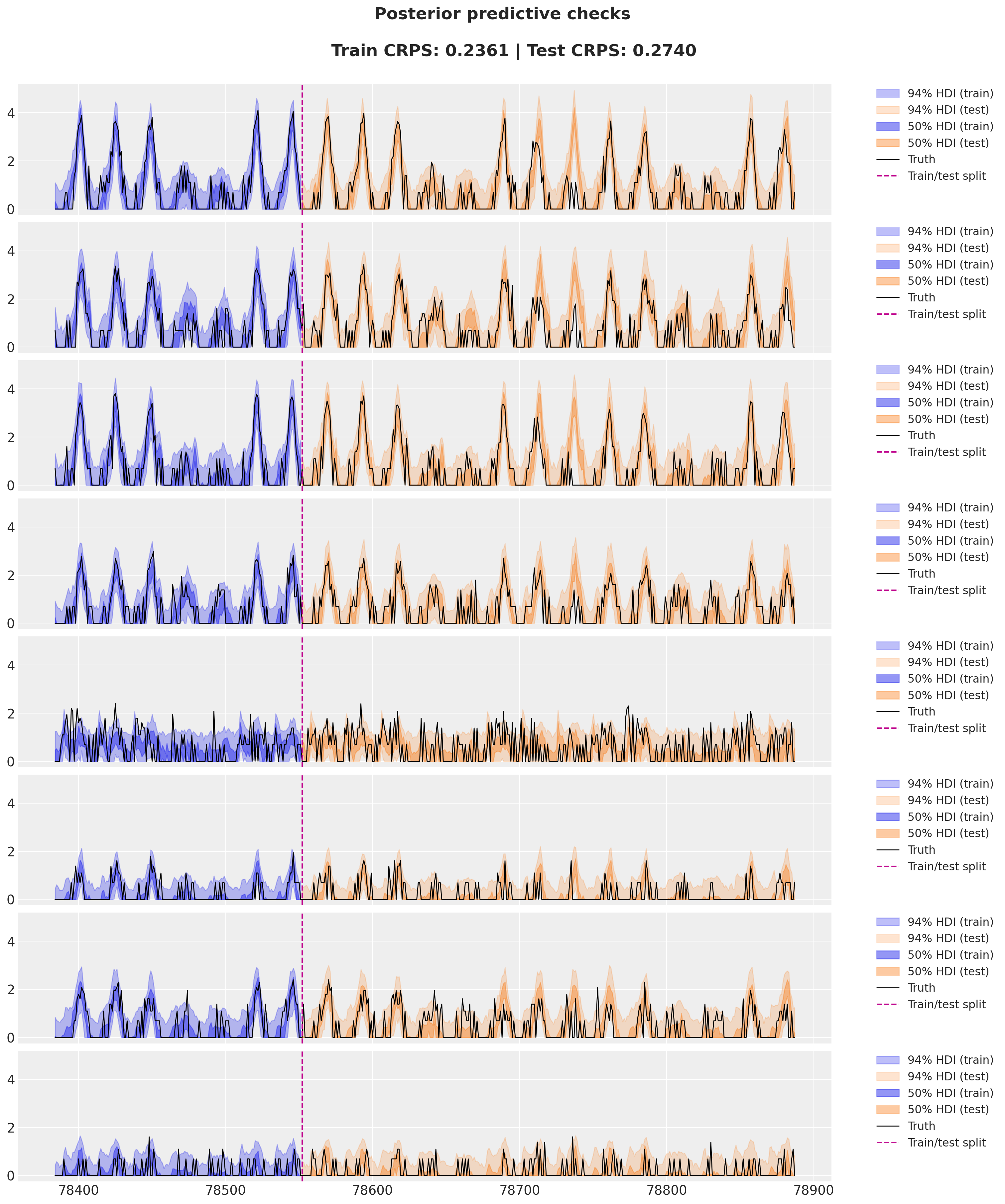
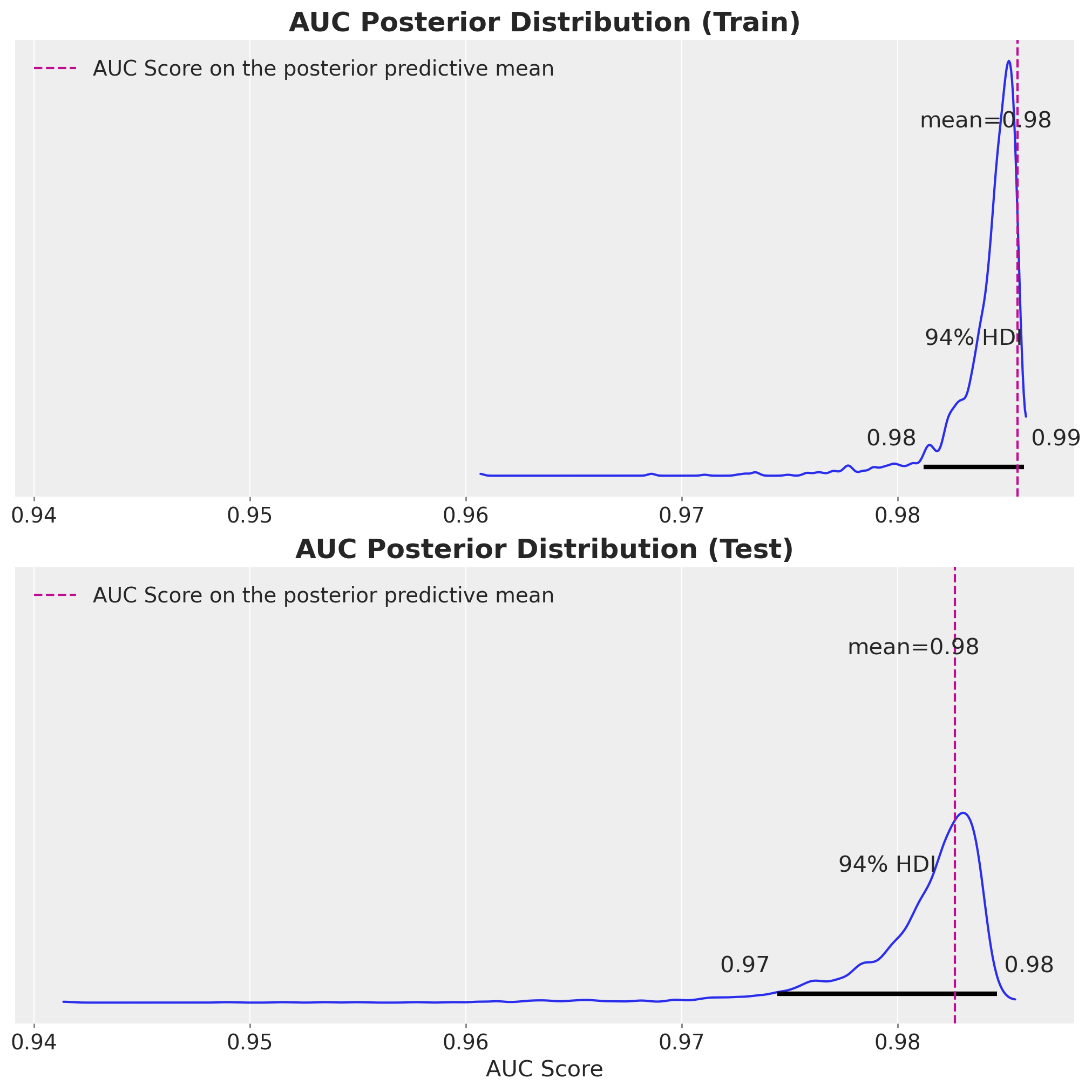
Model Evaluation: Posterior AUC
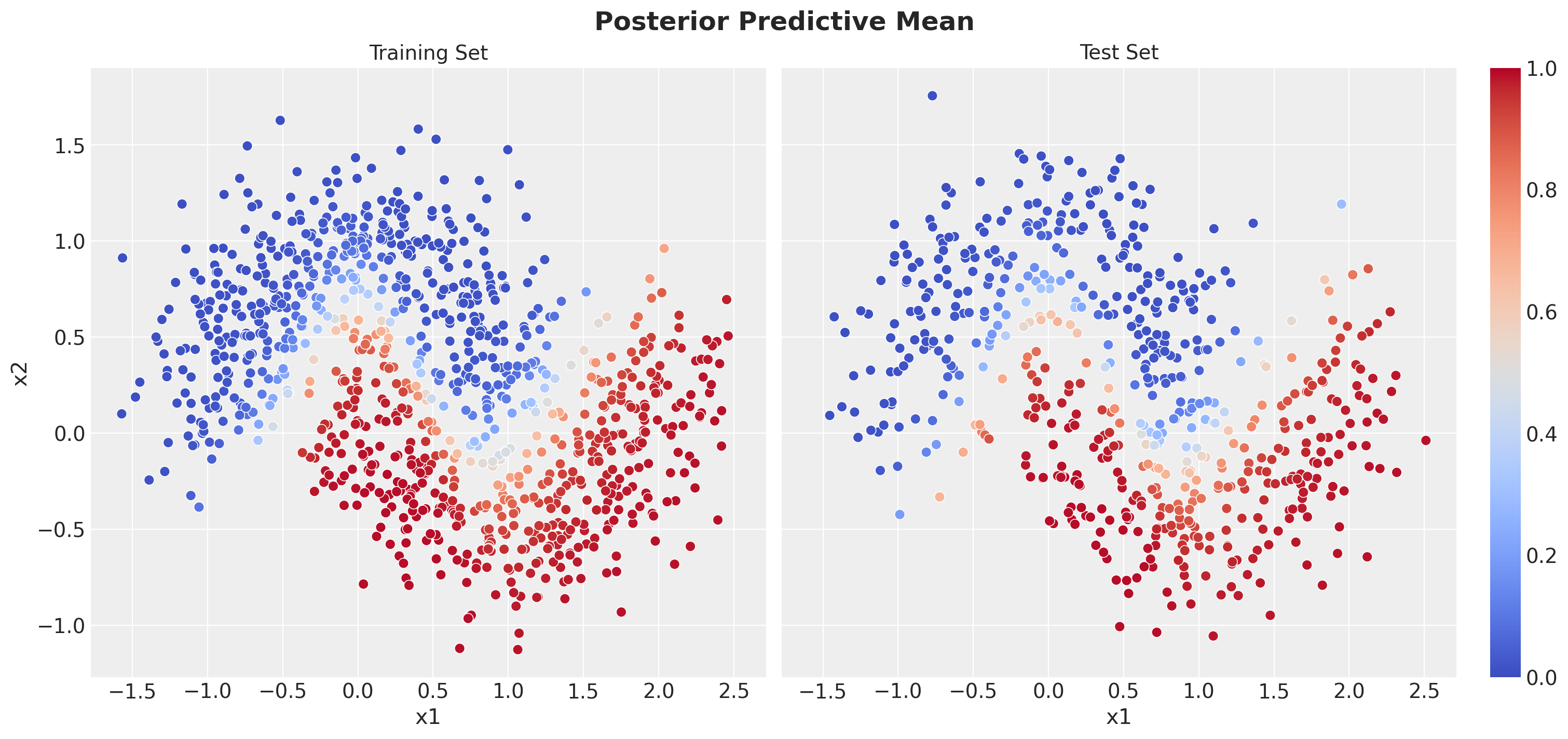
Posterior Predictive Mean
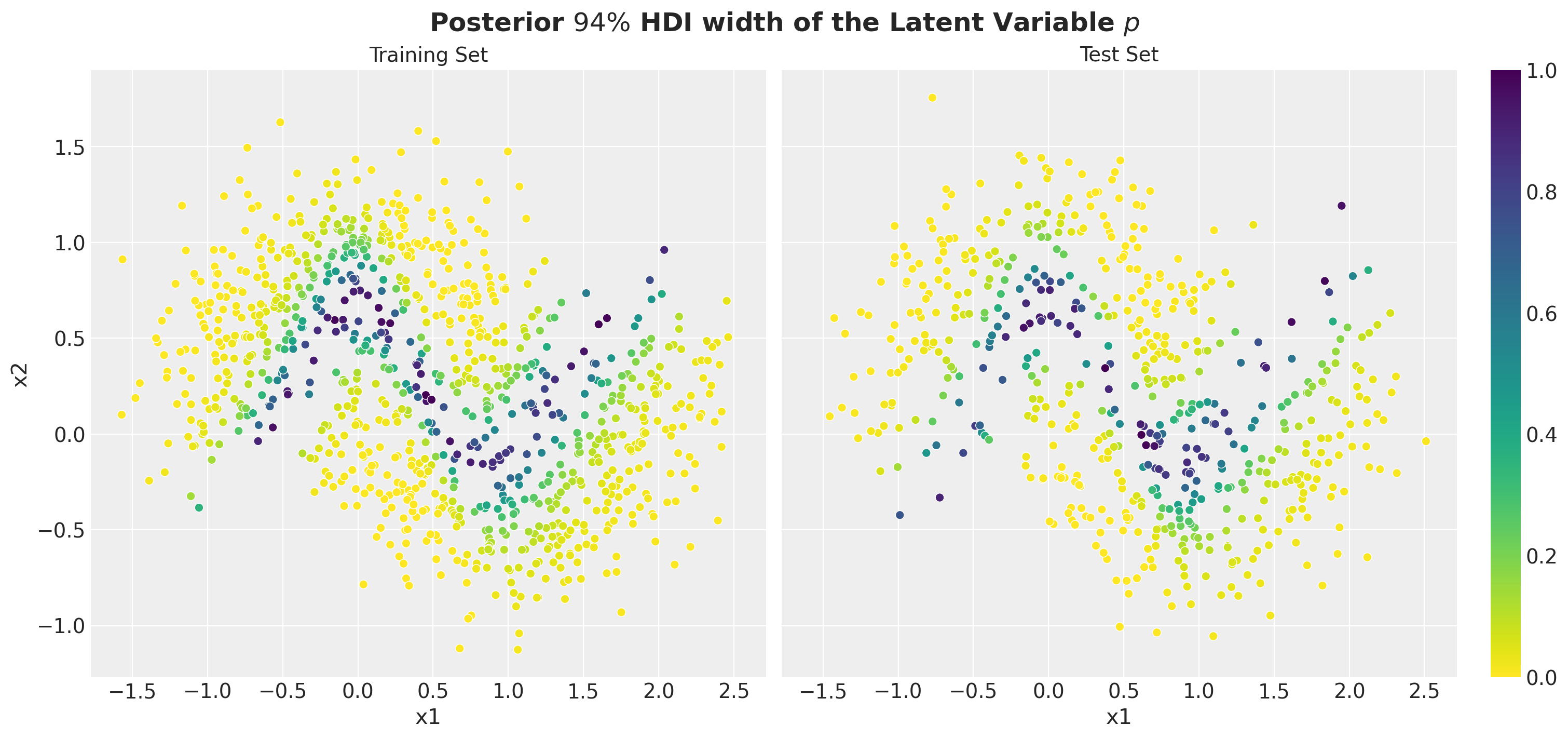
Posterior Predictive Uncertainty
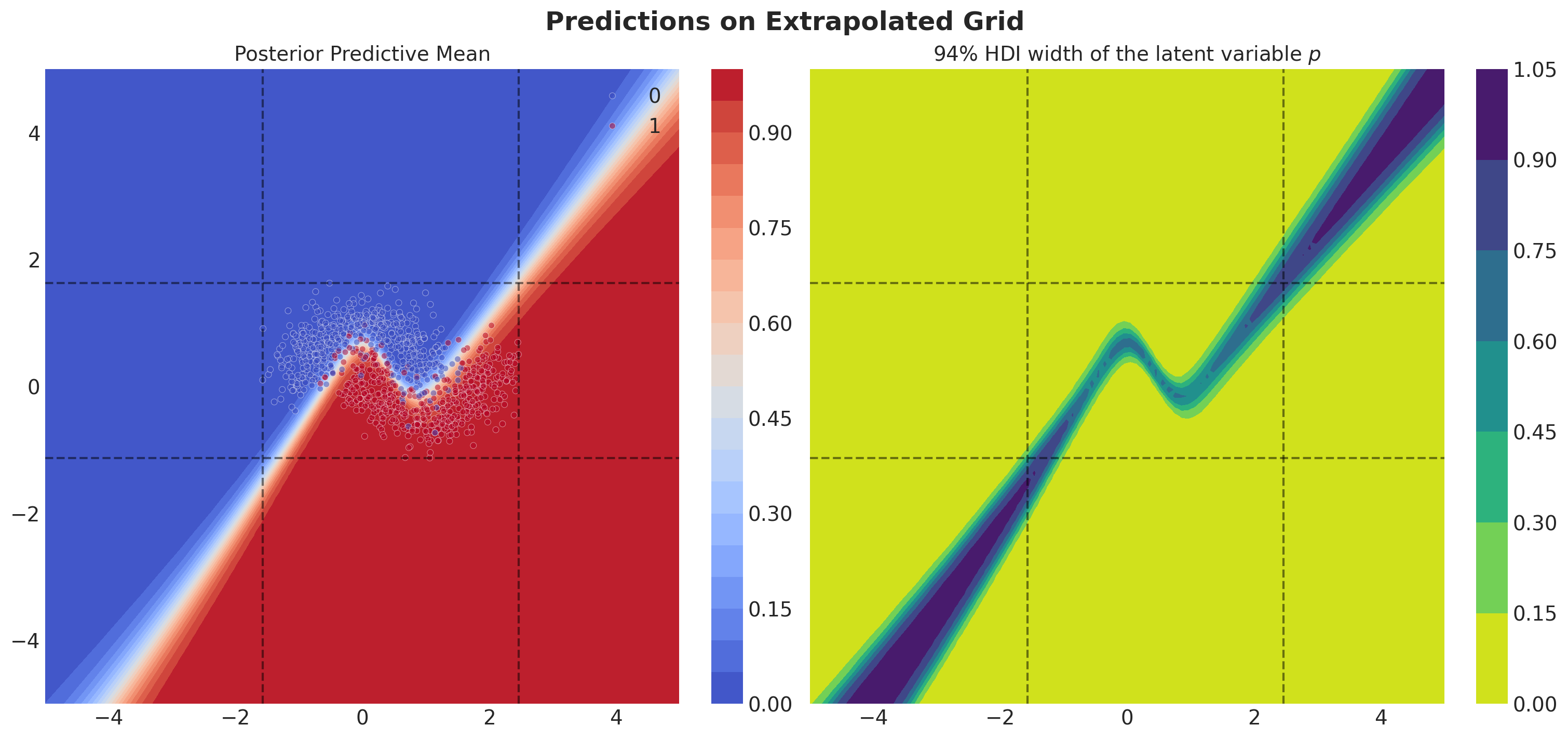
Predictions Outside Training Data
Tips & Tricks
When to Use SVI vs MCMC?
Use SVI when:
- You have large datasets
- You need fast inference for production systems
- You can accept approximate (vs exact) posterior inference
Use MCMC when:
- You have small-medium datasets
- You need precise posterior samples
- You have time for longer computation
Tips & Tricks
Tips and Tricks: SVI in Production
- Using GPUs for training can significantly speed up the training process.
- Using mini-batches can help with memory issues and speed up the training process.
- Using a good learning rate scheduler can help with the training process.
- Computing posterior predictive samples can be batched using
Predictivefrom NumPyro (you might need to do it by hand, but is quite straightforward). This batching can help memory issues.
Other Inference Methods for Bayesian Models
It is important to note that there are other inference methods for Bayesian models:
- See all the Markov Chain Monte Carlo (MCMC) methods available in NumPyro
- See some more custom ones from
BlackjaxorFlowJAX. - All of them can be integrated with NumPyro, see the example notebook NumPyro Integration with Other Libraries.
References 📚
Pyro Tutorials
- SVI Part I: An Introduction to Stochastic Variational Inference in Pyro
- SVI Part II: Conditional Independence, Subsampling, and Amortization
- SVI Part III: ELBO Gradient Estimators
- SVI Part IV: Tips and Tricks
- NumPyro Documentation
Papers
Videos
Thank you!
juanitorduz.github.io